ABSTRACT
Aim/Background
Arabica coffee (Coffea arabica) is the most consumed coffee in the world. As a result, it is susceptible to imitation Arabica coffee made with corn syrup and other inexpensive components (Zea mays). This study detects the corn Zein gene in Arabica coffee powder on the market with the Gel-based PCR method.
Materials and Methods
In this study, the samples of research were split into two groups. Samples are separated into two categories: test and control. Controls include unprocessed corn (Zea mays) and coffee beans (Coffea arabica). There are now four distinct varieties of Arabica coffee powder available in the Indonesian market (Coffee arabica). In this study, the simplex PCR method with the primer pair is the Zein gene, and the control primer pair is the ClpP gene as an Arabica coffee gene.
Results
The results of this study are that the four test samples show a band parallels to the size of 87 bp (Zein gene band).
Conclusion
In conclusion, all test samples (Arabica coffee powder on the market) are suspected of containing the Zein gene, the corn gene.
INTRODUCTION
Arabica coffee (Coffea arabica) is the world’s most popular and highest-quality coffee. According to,1 Indonesia is the 4th largest coffee-producing country globally. So Indonesian Arabica coffee producers are required to maintain the quality of these products. But on the other hand, the adulteration of Arabica coffee is vulnerable in production, which maximizes profits by reducing production costs compared to maintaining product quality. According to the study, several other ingredients are cheaper and commonly used as adulterants in coffee, including soybeans, wheat,2–4 corn, rice,5 and husks.6 Because Indonesia is the largest corn producer in Southeast Asia,7 it is possible to counterfeit Arabica coffee products using corn.
Previous research has conducted physical, chemical, and biological approaches to detecting adulterants in coffee. In the physical method, there are several detection techniques used, such as infrared,6 digital image processing,8 Mass spectroscopy,9 thermal lenses,10 photothermal,11 and 1H NMR spectroscopy.12 In the spectrometry method, infrared has a practical advantage because it is fast and does not require prior sample preparation. However, the spectrometry method also has the disadvantage of requiring complex statistical analysis and the need for a particular expertise. And chemical methods include using HPLC-HPAEC-PAD,13 HPLC-UV,14 HPLC with fluorescence,15 and SPME-GC-MS.16 The advantage of the chromatography method based on chemical screening carried out on the sample allows the identification of potential adulterant markers. However, the chromatography method is expensive and requires particular expertise.17
Another alternative method for detecting adulterants in coffee besides physical and chemical methods is the biological method with a molecular approach, namely the PCR technique.5 PCR is an alternative technique that is sensitive and specific to detect the adulteration of a food product.17 PCR is a DNA-based technique; target marker gene information is needed to identify the target’s presence in the sample. The Zein gene is a gene that is only found in corn.18 Therefore this study aims to detect the gene marker Zein corn (Zea mays) in coffee powder (Coffea arabica) on the market with the Gel-Based PCR method.
MATERIALS AND METHODS
Samples
In this study, there were two groups of samples: control samples, which included raw corn (Zea mays) and raw coffee beans (Coffea arabica), and test samples, which included four commercially available samples of Arabica coffee powder (Coffea arabica). These samples are purchased at the local market directly and online in Indonesia.
DNA Extraction
DNA extraction from samples of raw coffee beans and raw corn is using Wizard® Genomic DNA Purification KIT. The extraction from four samples of Arabica coffee powder on the market using the DNeasy mericon Food Kit. The results from DNA extraction are in the form of liquid containing pure DNA stored at -20°C until analysis.
DNA analysis with Gel-Based PCR method
Selection of oligonucleotide primer
There are several stages for primer selection: the literature study stage, the primer evaluation, and the primer sequence. The literature study in this research refers to one of the journals, selected primer, then evaluated by entering the primer information obtained into the NCBI BLAST tool (online). The evaluation stage by Snap Gene software after that from the selected primer sequence is sought information about the primer length, % GC, and melting Temperature (Tm), which is then analyzed and then made a primer order.
DNA amplification
5 µL of DNA was amplified in 1X buffer, 3.5 μM MgCl2, 10 μM dNTPs, 0.25 U/µL Taq polymerase, and 0.2 μM primers with a total volume of 50 μL. The settings on the Thermal PCR are 95°C for 5 min and 94°C for 50 sec (denaturation), 58°C for 1 min (annealing), 72°C for 50 sec, and 72°C for 5 min (elongation) with a total of 40 cycles for those using the primer of Zein gene (corn). Likewise, the PCR Thermal setting using ClpP (coffee) primer is the same as the setting on the corn primer, but the difference is from the annealing temperature of 50°C for 1 min.5
Gel-Agarose electrophoresis
The amplicon was electrophoresed on 2% gel containing 0.5 µL DiamondTM Nucleic Acid Dye and visualized using UV Transilluminator.
Data analysis
The visualization results of agarose gel electrophoresis showed the formation of a band at size 87 bp as the Zein (corn) gene and a band at length 114 bp as the ClpP gene (Arabica coffee). Control samples consisted of raw Arabica coffee beans and raw corn to confirm both bands. Therefore, in this study, the band’s reading is based on the length of the amplicon (bp) and compared with the control sample.
RESULTS AND DISCUSSION
Evaluation of DNA extraction
Analysis of the electrophoresis of the agarose gel that can be seen in Figure 1 shows that in the test sample (4 samples of the Arabica coffee powder are traded), the band obtained has a dark color. This can be due to the DNA extraction method and heating on the Arabica coffee powder processing process affecting the quality of the DNA produced. As the journal says19 that DNA extraction is one of the factors affecting the success of PCR-based methods, the journal explains that heat treatment involves the quality of DNA extracted from foodstuffs, as it can shorten the quality length of mitochondrial and nuclear DNA fragments.
Evaluation of DNA amplification
The primer pair selection in the study was derived from a literature study referring to one journal,5 so selected two genes to be used in the study of ClpP and Zein proteins. The two genes are then evaluated by entering information from the literature study results into the NCBI BLAST (online) tool; this stage aims to analyze the similarity of sequences with other species. This was followed by an evaluation using SnapGene software to ensure the primary sequence matched the sequences obtained. The selected primer sequence can be seen in Table 1 And from the primer pair chosen is sought information about the primer length, % GC, and melting Temperature (Tm), which is then analyzed. This primary analysis aims to determine the annealing temperature suitable for use in DNA amplification.
Result of the gel electrophoresis
The electrophoresis indicated by Figure 1 is an amplification of PCR in Simplex (single), with the primer used being the Zein primer, as listed in Table 1. Gen Zein is a gene only found in corn (18), so the primer designed from the Zein gene sequence is called a specific primer to detect the presence of corn on a product.
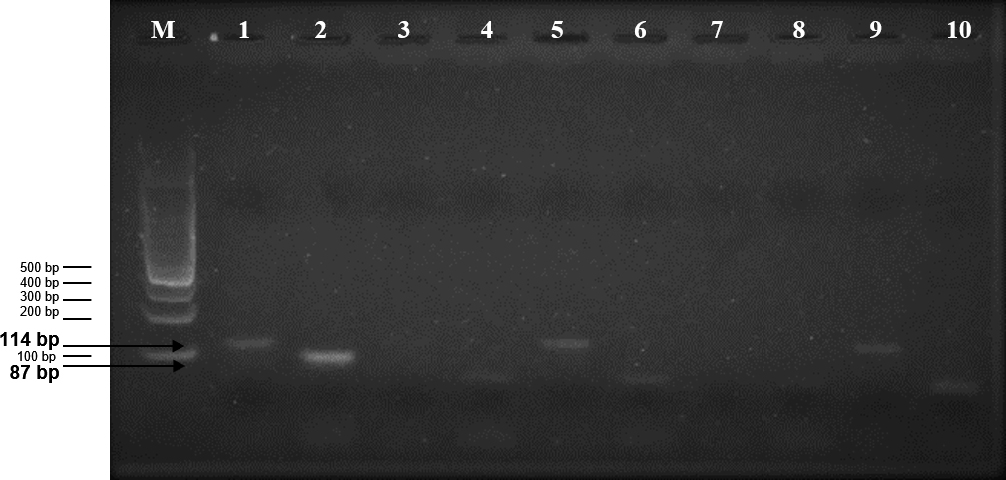
Figure 1:
Visualization of electrophoresis gel agarose detection of corn in Arabica coffee powder.
| Organisms | Gene | Name | Sequence 5’-3’ | Amplicon length (bp) | Reference |
|---|---|---|---|---|---|
| Corn | Zein protein | Zeina1-F | TGG CCA GCT AGC TAC AAC AAA CCG | 87 | 5 |
| Zeina1-R | GCG GGG TTA GCC GAA AAC TGCT | ||||
| Coffee | ClpP | Café1-F | TTC CGA AGT CCT GGA GAG | 114 | |
| Café1-R | CGG AGG ATA TCT CAA TCG |
Primer sequence for the detection of corn and Arabica coffee.
Description
M: 100 bp DNA Ladder.
1: Raw Arabica coffee beans with ClpP primer. 2: Raw corn with Zein primer.
3: Sample A of Arabica coffee powder on the market with ClpP primer.
4: Sample A of Arabica coffee powder on the market with Zein primer.
5: Sample B of Arabica coffee powder on the market with ClpP primer.
6: Sample B of Arabica coffee powder on the market with Zein primer.
7: Sample C of Arabica coffee powder on the market with ClpP primer.
8: Sample C of Arabica coffee powder on the market with Zein primer.
9: Sample D of Arabica coffee powder on the market with ClpP primer.
10: Sample D of Arabica coffee powder on the market with Zein primer.
Sample control: 1 and 2.
Test samples: 3, 4, 5, 6, 7, 8.9 and 10.
In this test, the primer pair (Zeina1-F and Zeina1-R) used has a size amplicon of 87 bp. The band with the amplicon size will provide information that the band indicates the existence of the gene Zein on the sample. In addition to the Zein gene as the primary gene detected, ClpP genes are also used to represent specific genes for Arabica coffee. The primer pair of the ClpP gene tested will show the amplicon of 114 bp as listed in Table 1 ClpP gene function in this test is a control that the sample used contains Arabica coffee.
The data analysis of the simplex amplification results in the electrophoresis gel-agarose on the test sample (Arabica coffee powder on the market) indicates a band parallels to the 87 bp or the parallel control sample, therefore the suspected zein gene in all test samples (Figure 1). All test samples contain ingredients other than Arabica coffee corn.
But the simplex amplification result in the test samples against the presence of Arabica coffee shows different results. As can be seen in Figure 1, not all test samples 3, 5, 7, and 9 indicate the presence of the Band of the primer gene ClpP because test samples 5 and 9 are suspected of showing a band that is parallel to the 114 bp or the control sample 1, while the test samples 3 and 7 are suspected not indicate the presence of the band gene ClpP, where Arabica coffee is the main ingredient that should be present in all of these test samples.
CONCLUSION
The results of this study show that the Gel-Based PCR method can be used to find the Zein corn (Zea mays) gene in commercial Arabica coffee powder (Coffea arabica). The gel electrophoresis results show that the Zein gene is present in all test samples (commercial Arabica coffee powder), which means that the four test samples are likely to contain another corn ingredient. As for the four test samples, only test samples of No. 5 and 9 were supposed to have Arabica coffee. And for further development of this research, it is necessary to perform more accurate quantifying validation.
References
- . Coffee: world markets and trade. Coffee World Mark Trade. 2019:1-9. [Google Scholar]

- Oliveira RCS, Oliveira LS, Franca AS, Augusti R. Evaluation of the potential of SPME-GC-MS and chemometrics to detect adulteration of ground roasted coffee with roasted barley. J Food Compos Anal.. 2009;22(3):257-61. [CrossRef] | [Google Scholar]

- Pauli ED, Barbieri F, Garcia PS, Madeira TB, Acquaro VR, Scarminio IS, et al. Detection of ground roasted coffee adulteration with roasted soybean and wheat. Food Res Int. 2014;61:112-9. [CrossRef] | [Google Scholar]

- Nogueira T, Do Lago CL. Detection of adulterations in processed coffee with cereals and coffee husks using capillary zone electrophoresis. J Sep Sci. 2009;32(20):3507-11. [PubMed] | [CrossRef] | [Google Scholar]

- Ferreira T, Farah A, Oliveira TC, Lima IS, Vitório F, Oliveira EMM, et al. Using Real-Time PCR as a tool for monitoring the authenticity of commercial coffees. Food Chem. 2016;199:433-8. [PubMed] | [CrossRef] | [Google Scholar]

- Reis N, Franca AS, Oliveira LS. Discrimination between roasted coffee, roasted corn and coffee husks by Diffuse Reflectance infrared Fourier Transform Spectroscopy. LWT Food Sci Technol. 2013;50(2):715-22. [CrossRef] | [Google Scholar]

- Asean ME. ‘Competitiveness of Indonesian’s Corn in Facing,’ no. Koesrianti. 2016 [CrossRef] | [Google Scholar]

- Sano EE, Assad ED, Cunha SAR, Correa TBS, Rodrigues HR. Quantifying adulteration in roast coffee powders by digital image processing. J Food Quality. 2003;26(2):123-34. [CrossRef] | [Google Scholar]

- Garrett R, Vaz BG, Hovell AMC. Eberlin, Rezende C.M.. J Agric Food Chem.. 2012;60(17):4253-8. [PubMed] | [CrossRef] | [Google Scholar]

- Fontes AS, Bento AC, Miranda LCM, Baesso ML. Thermal lens evaluation of the presence of adulterants in brewed coffee. Anal Sci. 2001;17:526-9. [PubMed] | [CrossRef] | [Google Scholar]

- Fontes AS, Bento AC, Baesso ML, Miranda LCM. Thermal lens and pH measurements in pure and adulterated brewed coffee. Instrum Sci Technol. 2006;34(1-2):163-81. [CrossRef] | [Google Scholar]

- de Moura Ribeiro MV, Boralle N, Redigolo Pezza H, Pezza L, Toci AT. Authenticity of roasted coffee using1H NMR spectroscopy. J Food Compos Anal.. 2017;57:24-30. [CrossRef] | [Google Scholar]

- Domingues DS, Pauli ED, de Abreu JE, Massura FW, Cristiano V, Santos MJ, et al. Detection of roasted and ground coffee adulteration by HPLC by amperometric and by post-column derivatization UV-vis detection. Food Chem. 2014;146:353-62. [PubMed] | [CrossRef] | [Google Scholar]

- Núñez N, Collado X, Martínez C, Saurina J, Núñez O. Authentication of the origin, variety and roasting degree of coffee samples by non-targeted HPLC-UV fingerprinting and chemometrics. Application to the detection and quantitation of adulterated coffee samples. Foods. 2020;9(3):378 [PubMed] | [CrossRef] | [Google Scholar]

- Alves RC, Casal S, Oliveira MBPP. Tocopherols in espresso coffee: analytical method development and validation. Food Chem. 2009;115(4):1549-55. [CrossRef] | [Google Scholar]

- Toci AT, Farah A.. Volatile fingerprint of Brazilian defective coffee seeds: corroboration of potential marker compounds and identification of new low quality indicators. Food Chem. 2014;153:298-314. [PubMed] | [CrossRef] | [Google Scholar]

- Habza-Kowalska E, Grela M, Gryzińska M, Listos P.. Molecular techniques for detecting food adulteration. Med Weter. 2019;75(7):404-9. [CrossRef] | [Google Scholar]

- Anderson TJ, Lamsal BP. Review: Zein extraction from corn, corn products, and coproducts and modifications for various applications: a review. Cereal Chemistry Journal. 2011;88(2):159-73. [CrossRef] | [Google Scholar]

- Liao J, Liu YF, Ku T, Liu MH, Huang Y.. Qualitative and quantitative identification of adulteration of milk powder using DNA extracted with a novel method. J Dairy Sci. 2017;100(3):1657-63. [PubMed] | [CrossRef] | [Google Scholar]

