ABSTRACT
The new coronavirus has spread to more than 200 nations since it first appeared in China in December 2019. Worldwide, there have been more than 58.3 billion confirmed cases of the coronavirus and more than 64.2 lakh fatalities as of August 2022. In India, more than 4.41 billion confirmed cases and more than 5.27 lakh fatalities have occurred. The outbreak of coronavirus 2019 (COVID-19) was caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In India, the outbreak led to the imposition of a nationwide lockdown, which resulted in a sharp decrease in the number of new confirmed cases. However, in recent months, there has been an increase in new COVID-19 cases in India, with a number of new variants of the virus being detected. The variants found in India are Alpha, Beta, Gamma, Delta, Kappa, Omicron, and B.1.618 (Triple mutant). Although the symptoms of the virus closely align with the common cold, during severe infection, one may experience loss of smell and taste sensation and long-term infection can lead to post-COVID complications. The Indian government has implemented a number of treatment strategies in an effort to control the spread of the disease and reduce the number of fatalities. As the pandemic situation continues to evolve, there is an urgent need to understand the epidemiology of the disease and the characteristics of the virus. In this review, we provide a summary of our current understanding of the novel coronavirus, the mutant novel strains that are prevalent in India, the epidemiology of COVID-19, and the control measures that have been put in place in India and their effectiveness in halting the disease’s spread.
INTRODUCTION
The novel Coronavirus causes severe acute respiratory syndrome-2 which belongs to the family Coronaviridae. The symptoms are similar to the influenza virus which belongs to the family Orthomyxoviridae their structure, incubation time, and severity vary though they share the same form of transmission through infected droplets, fomites, and person-to-person contact. The Novel Coronavirus has caused a global pandemic condition, resulting in health crises across 213 countries around the globe. According to official reports, the USA recorded the maximum number of cases and deaths. The first and second waves have also presented a challenge to India’s healthcare sector because of the daily increase in cases and death rates. Many countries imposed strict rules to control the increasing number of cases. In spite of following many preventive measures, the infection is still difficult to control due to its high virulence. The high virulence is attributed to its constantly mutating spike glycoprotein. The World Health Organization designated COVID-19 as a Public Health Emergency of International Concern on January 30, 2020, and on March 11, 2020, it deemed the situation to be pandemic. This worldwide catastrophe has allowed people to try out new initiatives such as following social distancing, remote working, and the use of technology in education and health.
The novel Coronavirus is composed of the RNA genome which is an omnipresent cellular parasite that generally infects mammals. They belong to the kingdom Orthornavirae, Phylum Pisuviricota, order Nidovirales, family Coronaviridae and genus betacoronavirus. SARS-CoV-2 are enveloped positive-sense single-stranded RNA virus with a genome of 28-33 kb in size considered to be the largest RNA virus. There are 4 genera of coronaviruses namely Alpha, Beta, Gamma, and Delta of which alpha and beta coronaviruses got more attention due to their potential to cross animal-human barriers. SARS-CoV, MERS-CoV, and SARS-CoV-2 belong to the Beta coronavirus genera which cause mild symptoms like the common cold, fever, fatigue to severe symptoms like difficulty in breathing, and chest pain which leads to mortality. The first SARS-CoV-2 case in India was reported on 27th January 2020 in Kerala. Then the cases spread rapidly across various states in India and started affecting thousands of people daily. India faced a challenge from the second SARS-CoV-2 wave, which led to higher mortality rates because of a shortage of oxygen supplies for the patients leading to a peak in the fatality rate.
EMERGENCE OF NOVEL CORONAVIRUS
SARS-CoV-2 belongs to β-coronavirus, was detected in Wuhan, China in December 2019, which swiftly spread to many countries globally and led to a severe pandemic.1 By January 2021, 97 million people were affected and 1.4 million deaths were reported globally.2 The novel Coronavirus (nCoV), which first appeared in December 2019, was found in the Huanan seafood market in Wuhan State, Hubei Province, China, where livestock was traded. On December 12, 2019, an unidentified case of pneumonia was documented, and subsequent laboratory testing ruled out any presence of influenza or other coronaviruses. On January 7, 2020, Chinese authorities confirmed that they had identified a novel coronavirus (nCoV). This virus was named Novel Coronavirus by WHO on January 12 2020 and COVID-19 on 11 February 2020.3 The infection was transmitted from animal to human. The increase in the number of cases in Wuhan city and the secondary spread of the disease was from human to human. The identification of the genome sequence of SARS-CoV-2 (Wuhan-Human_1) was first released by China on 10th January 2020.4
SARS-CoV-2 and Bat CoV RaTG13 sequences were genetically similar suggesting that bats might be natural reservoirs for SARS-CoV-2. Researchers also found that pangolins might be intermediate hosts as their sequence and SARS-CoV-2 sequence are similar. Bats transmitted the virus to humans through intermediate hosts and this leads to human-human transmission or communal spread. SARS-CoV-2 is disseminated through Respiratory droplets while coughing or sneezing.5 More than 7 strains of human coronaviruses (hCoV’s) were identified which includes 3 Beta genera CoVs namely SARS-CoV, MERS-CoV, and SARS-CoV-2, hCoV-HKU1, and hCoV-OC43. Two Alpha genera CoVs namely, hCoV-NL63 and hCoV-229E. Except for SARS-CoV, MERS-CoV, and SARS-CoV-2, all the other strains are already circulating among humans They either don’t cause any symptoms or may cause a common cold (5-30%).5
SEVERE ACUTE RESPIRATORY SYNDROME (SARS)
SARS-CoV is enveloped by single-stranded, positive-sense, and highly diverse RNA viruses.5–7 The endemic outbreak was first identified on 16th November 2002 in Foshan, Guangdong, China. This caused a global epidemic condition involving 29 different countries. Over 8000 people were infected with a mortality of about 774 deaths reported until July 2003 with a case fatality ratio of 9.6%. The β-coronavirus, was the causative agent of severe acute respiratory syndrome (SARS) and its transmission was from palm civet.5–9 The genome of the SARS-CoV like other RNA viruses, Coronavirus also mutates rapidly due to the error-prone nature of RNA polymerases and their short replicative life cycles.10
STRUCTURE OF SARS-CoV
The 5′ methylated caps and 3′ polyA tails that make up the typical genomic structure of beta coronaviruses are present in the SARS-CoV, MERS-CoV, and SARS-CoV-2. Spike protein, envelope protein, membrane protein, and nucleocapsid protein are structural proteins that are essential for the viral life cycle and are encoded by the 3′-terminal regions. The non-structural protein-coding region of the 5′-terminus, which makes up two-thirds of the sequence, contains important genes for viral replication.10
The structural proteins as shown in Figure 1 specifically spike (S), membrane (M), envelope I, and nucleocapsid (N) proteins are responsible for the attachment to the host cells of humans, mammals, and avian species resulting in excess of the infectious prions causing respiratory and enteric diseases.10–11 The process of replication in the host cells is a cascade of events that are encoded by the genes present in the S region which enables the viral invasion into the host cell involving the cytoplasmic domain (CP), fusion protein (FP), heptad repeats (HP), receptor binding site (RBD), receptor binding motif (RBM), signal peptide (SP), and the transmembrane protein I which acts as a major virulence factor in Human Coronavirus (HcoVs). Angiotensin-converting enzyme 2 (ACE2) receptors which are the membrane-bound host cell receptors have a vast biodistribution, including respiratory tract, gastrointestinal tract, heart, kidney, and olfactory neuroepithelium, resulting in broad cellular and tissue tropisms of SARS-CoV.12
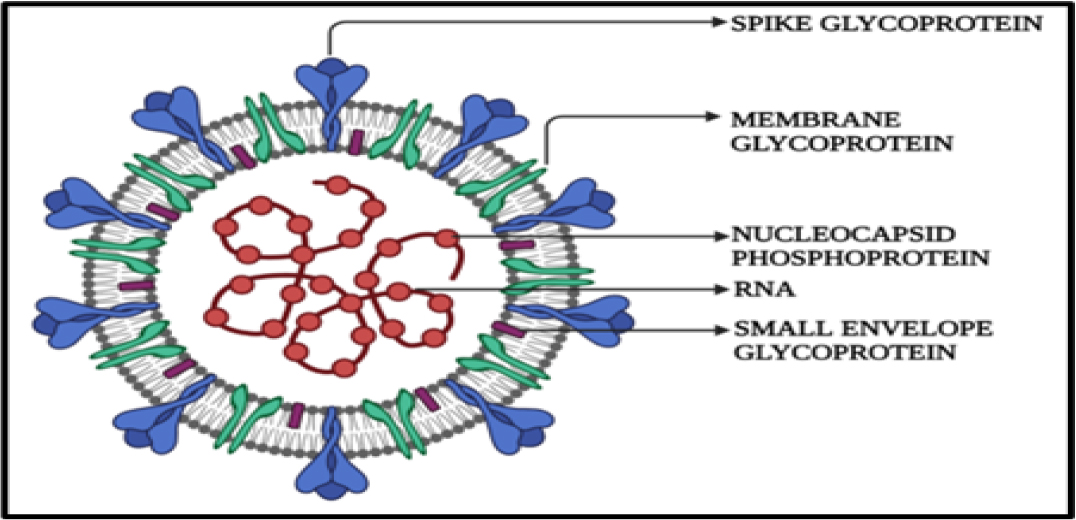
Figure 1:
Structure of SARS-CoV.
STRUCTURE OF SARS-CoV-2
A class I viral fusion protein titled as spike protein, as depicted in Figure 2, adopts a metastable prefusion conformation following translation and assembles into trimers that resemble club-shaped spikes together with the CoV membrane surface. The spike protein is made up of three parts: the ectodomain, a single-pass transmembrane anchor, and a brief intracellular tail. The S1 receptor-binding subunit, which participates in viral attachment, and the S2 membrane-fusion subunit, which promotes virus-cell fusion, are further subdivided into the ectodomain. The S1 will bind to a suitable receptor on the membrane of the host cell to enable viral attachment, and the S2 will fuse to both the host and viral membranes to enable viral genomes to enter the host cells.13
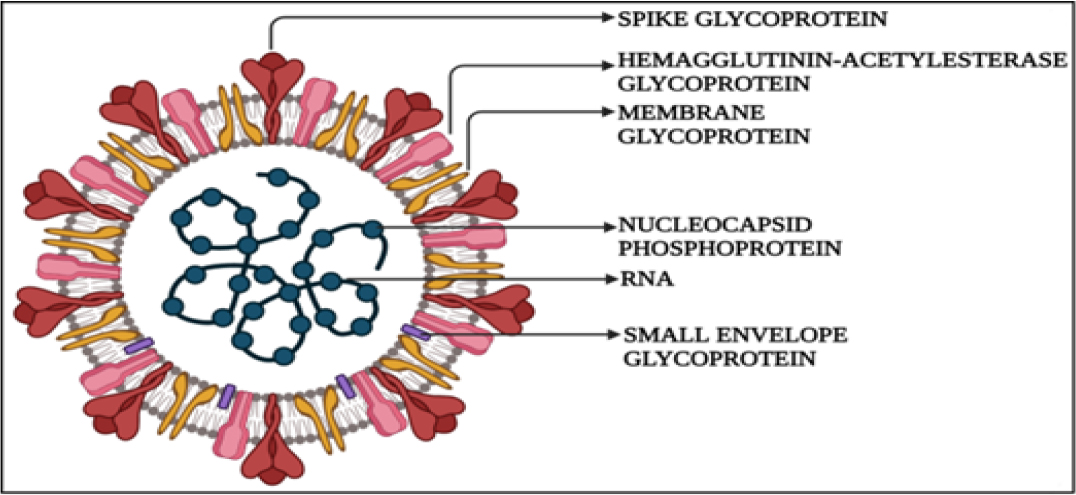
Figure 2:
Structure of SARS-CoV2
The S protein has a transmembrane I domain that is anchored, an extracellular N-terminus, and a brief intracellular C-terminal region. Polysaccharide molecules are coated on the spikes, helping them to blend in and evade the host immune system. The SARS-CoV-2 spike protein comprises two subunits the S1 subunit and the S2 subunits, with a total length of 1273 aa and consists of a signal peptide located at the N-terminus.13 The N-terminal domain and a receptor-binding domain are present in the S1 subunit, and the S2 subunit comprises the fusion peptide, heptapeptide repeat sequence, HR2, TM domain, and cytoplasm domain. The structural variation between the coronaviruses is that the SARS-CoV lacks the hemagglutinin-esterase protein which is present in SARS-CoV-2. MERS-CoV specifically binds to (Dipeptidyl-peptidase 4) DPP4 while SARS-CoV and SARS-CoV-2 bind to ACE2 receptors. Spike protein is the main component that binds all three viruses with the host receptor.
LIFE CYCLE OF SARS-CoV-2
The SARS COV-2 viruses undergo a lytic lifecycle as shown in Figure 3 where the attachment to the host membrane is facilitated by spike protein (S) of SARS-CoV-2. The receptor binding domain (RBD) of the viruses has a higher affinity for Hace-2 receptors which are present in the respiratory tract.14 Transmembrane Serine Protease 2 (TMPRSS2) is a cell surface protein that helps in the priming of S protein. The role of TMPRSS2 is to cleave S1 and S2 domains to activate S protein to prevent non-endosomal pathways. The fusion of spike proteins and ACE 2 receptors causes a conformational change in the spike protein aiding in the fusion of viral entry into the host cell through the process of endocytosis.14 The endocytosed virus is called a viral endosome is uncoated, and genomic positive sense RNA was released and transcribed by the host cell ribosome. ORF1a and ORF1b are the largest gene on the RNA of SARS-CoV-2 which undergoes translation. During this process, two polyproteins are synthesized namely PP1a and PP1b which are processed into individual mass structure proteins called non-structural proteins (nsps).13 These nsps form the viral replication and transcription complex. The positive RNA strand is converted into a negative RNA strand which is used either for replication of the positive RNA strand for the assembly of new virions or for the transcription of sub-genomic mRNAs which are translated to structural proteins such as S, M, E, N, and accessory proteins. The nucleocapsid protein (N) attaches to the genomic positive sense RNA to produce a nucleoprotein complex while other proteins such as S, E, and M enter the endoplasmic reticulum (ER). Both nucleoprotein complex and other structural proteins move towards the ER-Golgi Intermediate compartment where the virion assembly and maturation occur leading to the budding and release of a newly synthesized virus. The newly released virion infects a new cell thereby leading to disease progression.15
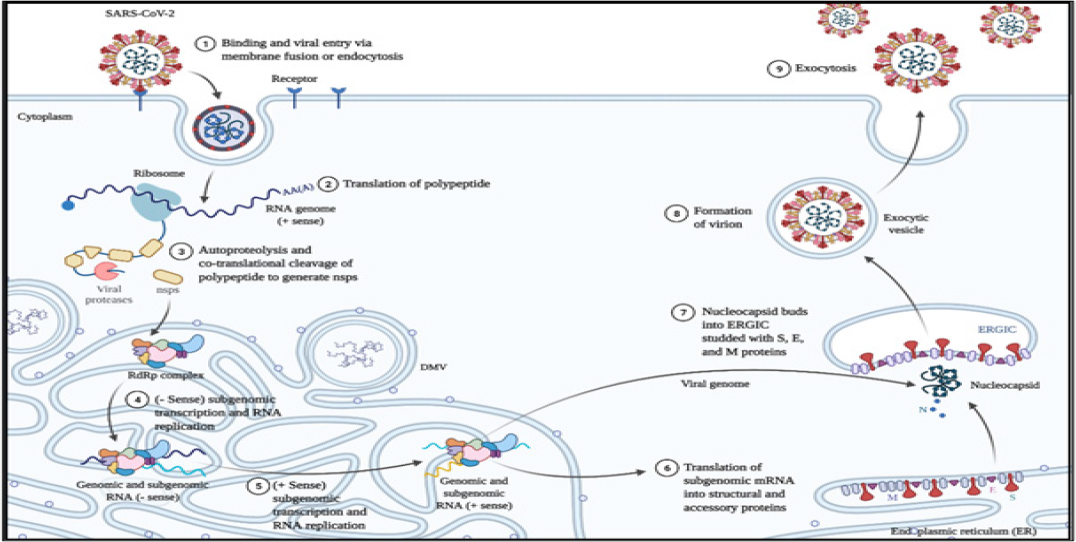
Figure 3:
Life Cycle of SARS-CoV2
SARS-COV-2 VARIANTS IN INDIA
SARS-COV-2 has undergone variations mainly due to RNA viruses. Hence, RNA-dependent RNA polymerases play an important role during viral nucleotide replication. These RNA polymerases are well known for their error-prone replication mechanism, which serves as the source for viral mutations, thereby leading to their evolution. The spike protein of SARS-CoV-2 has been shown to mutate rapidly, which may contribute to the virus’s ability to evade the immune system. The mechanism of spike protein mutation is not yet fully understood, but scientists have hypothesized that mutations are a consequence of random genetic drift. Based on an analysis of sequencing data archived in public repositories, the SARS-CoV-2’s genomic mutation rate in humans is estimated to be 0.8–2.38×10–3 nucleotide substitutions per site per year.16–17
The first genome sequence of SARS-CoV-2 became available in the Global initiative on sharing all influenza data (GISAID) consortium around January 10, 2020 (WH-Human or hCoV-19). India became the 5th country to isolate the viral genome from the first patient in Kerala.15 Over a period of time, the virus has undergone mutations two folds per month, leading to the emergence of multiple variants which have been detected. According to the COVID-19 Beacon database (CRL Viral Beacon), this mutation occurrence is 76.31% in the Indian population. The WHO has recommended using the Greek alphabet, namely alpha, beta, gamma, and delta, for naming the variants of SARS-CoV-2, which will be an informal terminology while discussing the details in the public domain. The SARS-CoV-2 variants as shown in Table 1 of concern that descends from this lineage were segregated and labelled according to their destination of occurrence. The overall distribution as shown in Table 1 of the SARS-CoV-2 variant in India as is as follows:
| Sl. No | Variant | Important spike protein mutation | Key characteristics of the mutant | Concern | Complications Post-covid | References |
|---|---|---|---|---|---|---|
| 1. | Alpha B.1.1.7 October 2020-November 2021 | N501Y D614G P681H | Single amino acid mutation. Receptor binding domain (RBD) of spike protein. | -Increased transmissibility. -Increased severity. | LUNG: Dyspnea Post covid diffuse lung disease Pulmonary embolism Respiratory muscle weakness Mucormycosis NEUROLOGICAL: Myalgic encephalitis Chemosensory disorders (smell and taste disturbances) Dysautonomia Cognitive impairment Sleep dysfunctions Neuromuscular disease Guillain-Barré syndrome Stroke Epilepsy NEPHROLOGICAL: Acute kidney injury Rapid progression of pre-existing chronic kidney disease Glomerular diseases Hypertension GASTRO-INTESTINAL: Dyspepsia Diarrhea Abdominal pain CARDIOVASCULAR: Increased cardiometabolic demands Myocardial fibrosis Left ventricular dysfunction Heart failure Arrhythmia Inappropriate sinus tachycardia Autonomic dysfunction | Kläser, Kerstin , et al. 2022 Loconsole, Daniela , et al. 2021 Vitiello, Antonio , et al. 2022 |
| 2. | Beta B.1.351 December 2020-June 2021 | K417N E484K N501Y D614G | Stronger affinity of the spike protein for the ACE2 receptor. | -Increased transmissibility. -Increased severity. -Possible reduction of vaccine effectiveness. | Kläser, Kerstin , et al. 2022 Loconsole, Daniela , et al. 2021 Vitiello, Antonio , et al. 2022 Cherian, Sarah , et al. 2021 | |
| 3. | Gamma P.1 March 2021-August 2021 | K417T E484K N501Y D614G | Stronger affinity of the spike protein for the ACE2 receptor. | -Increased transmissibility. -Increased severity. -Possible reduction of vaccine effectiveness. | Kläser, Kerstin , et al. 2022 Loconsole, Daniela , et al. 2021 Vitiello, Antonio , et al. 2022 Cherian, Sarah , et al. 2021 | |
| 4. | Delta B.1.617.2 November 2020-June 2022 | L452R D614G P681R | Mutation in the spike protein. | -Highly transmissible. -Highly severe. -Possible reduction of vaccine effectiveness. | Kläser, Kerstin , et al. 2022, Loconsole, Daniela , et al. 2021 Cherian, Sarah , et al. 2021 Vitiello, Antonio , et al. 2022 | |
| 5. | Kappa B.1.617.1 March 2020-May 2022 | E484Q L453R | Stronger affinity of the spike protein for the ACE2 receptor. | -Increased transmissibility. | Vitiello, Antonio , et al. 2022 Cherian, Sarah , et al. 2021 Singh, Jitendra , et al. 2021 | |
| 6. | B.1.618 October 2020-April 2021 | E484K D614G | Increased binding affinity of the spike protein for ACE2 receptor. | -Increased transmissibility. -Escape immune system. | Cherian, Sarah, et al. 2021 | |
| 7. | Omicron BA.4 BA.5 BA.2.75 December 2021-July 2022 | K417N E484K N501Y D614G L452R G446S R493Q | The mutation has increased binding affinity of the spike protein for ACE2 receptor. | -Increased transmissibility. -Possible reduction of vaccine effectiveness. -Escape immune system. | Sharma, Vineet , et al. 2022 Vitiello, Antonio , et al. 2022 |
Novel Corona Variants and their post COVID complications.
Due to their propensity for fast mutation and the severity of their means of transmission, the SARS-CoV-2 variations during each phase of the pandemic have been labelled as “variants of concern”. The SARS-CoV-2 spike protein variant D614G of the lineage B.1 bears mutation in S protein which is the most important stage of the infectivity as the S1 spike protein are involved in the binding to the host receptor through the receptor binding domain (RBD), which directly binds to the peptidase domain (PD) of angiotensin-converting enzyme 2 (ACE2) and the S2 is responsible for membrane fusion.
The Alpha variant B.1.1.7 was detected during the first wave in United Kingdom and was considered to be the most potent variant which had shown a significantly higher affinity for ACE2, hence was considered to be the dominant strain in circulation during the beginning of the pandemic. The Alpha variant B.1.1.7 had encountered non-synonymous mutations leading to mutants 501Y and N501Y which are reverse genomic mutations on a single gene and mutation of the spike protein. The Delta variant which was the major cause of the surge in India from Jan – March 2021 was considered to be the second wave which was 1000 folds more potent than the alpha variant. The clinical symptoms were more complex leading to common Influenza symptoms with further complications such as hearing impairment, clotting of blood vessels leading to tissue damage and gastrointestinal disorders.
The Kappa variant B.1.617 of SARS-CoV-2 variant isolated with two mutants – E484Q and L452R also called the double mutant virus. These double mutants were identified from Maharashtra, New Delhi, and Chandigarh. A new type of variant called B.1.618 is also called a triple mutant due to Variant Pseudo typed Lentiviruses were produced with SARS-CoV-2 B.1.618 Variant Spike an additional mutation E484K. These triple mutants are prevalent in Maharashtra, New Delhi, and West Bengal. These variants are capable of escaping the immune system thus evading the immune barriers and are also highly transmissible. Apart from these mutations, the spike proteins have undergone changes in the amino acid causing mutation of the spike protein. The more the number of mutations in the spike protein, the easier for the virus to gain access to the host cell since the spike protein binds to the host cell receptor.16–17
The Omicron variant B.1.1.529 of the SARS-CoV-2 variant was isolated in India on December 2, 2021. Since then, various subline-ages of this variant have been identified such as BA.2, BA.2.75, BA.4 and BA.5. While WHO has warned that BA.1.1.529 is spreading at an alarming rate, scientists have discovered that the variant has approximately 32 mutations in their spike proteins, the majority of which have been linked to antibody binding sites, implying that it would evade the immune system.18
The virus incubation period ranges from 2 to 10 days, however, the typical time between the onset of clinical symptoms and hospital admission is estimated to be between 3 and 5 days. Clinical symptoms that are frequently present include chronic fever, chills/rigor, myalgia, malaise, dry cough, headache, and dyspnea. Less frequently experienced symptoms include sore throat, rhinorrhea, nausea, vomiting, and diarrhoea.19 Patients affected showed abnormal chest X-ray, Patchy infiltrates, opacities, and areas of consolidation were the most noticeable radiographic findings. In 23% of the radiographs, multifocal lesions were visible. In lungs, CT scans indicated ground-glass opacities and unifocal or multifocal accumulation areas.20
STRATEGIC PREVENTION AND CONTROL
Standard operation protocol on Prevention and Control
Controlling the spread involves periodically washing hands with soap or an alcohol-based hand rub, cleaning surfaces with 70% ethanol alcohol, or wiping down surfaces with a 0.5 percent bleach solution. It was advised to practice respiratory hygiene, such as coughing or sneezing into a tissue, to avoid touching the eyes, nose, or mouth, and to dispose of contaminated tissue as soon as possible. The only practices that have been found to be effective in preventing the transmission of COVID-19 are the use of a surgical mask by individuals, keeping social distance in public settings, and working from home to avoid crowds.
Wearing mask
To avoid SARS-CoV-2 transmission, it was recommended that people wear masks, specifically non-valved multi-layer fabric masks. Masks are specifically designed to minimize the emission of virus-laden droplets, which is particularly important for asymptomatic or pre-symptomatic infected individuals who are unaware of their infectiousness to others and this kind of individuals accounts for more than 50% of transmission.21 It is mandated to dispose of the mask properly after use and not reuse it.
Quarantine and Isolation
When persons who are healthy but have been exposed to an infectious agent or disease are placed under a quarantine, the main objective is to track symptoms and identify cases as soon as possible.22 Quarantine helps to slow down the rate of infection. Quarantine has proved to be effective in China because the government has extensive electronic surveillance and physical control over its population which is difficult to achieve in India. Seclusion of an infected person from others to prevent the spread of infection to others is considered an important method in coping with infectious diseases like COVID-19, which has a high rate of spread from infected droplets.22
Treatment strategies
So far there are no specific medicines discovered to treat COVID-19. The most common treatments are symptomatic management and supportive care. However, patients are treated based on the severity of the infection. In case of mild infection with SpO2 levels between 94-97%, symptoms such as fever, sore throat and upper respiratory tract infection, patients were asked to be in-home isolation for 14 days from the date of testing positive or onset of symptoms.23 Medicines such as antipyretics and antibiotics are prescribed by the doctor. SpO2 levels should be periodically monitored. The patients were asked to wear a mask and isolate them from people in and outside their residences all the time.
In case of moderate infection with oxygen levels between 90-94% with symptoms of pneumonia, patients are admitted to a hospital. The vital signs of patients are routinely monitored in hospitals. If the patient has respiratory issues, oxygen support is provided. Anti-inflammatory medications such as glucocorticoids and toxilizumab will be provided to minimize excessive inflammation in order to prevent future harm to the lungs from inflammation. Antiviral drugs such as Remedisiver, Chloroquine, Hydroxychloroquine, Lopinavir will be prescribed if necessary.24
Anticoagulant therapy
The risk of Venous Thromboembolism (VTE) in people with SARS-CoV-2 is predicted by the D-dimer level, prothrombin time, fibrinogen level, and platelet count. Increases in fibrin, fibrin breakdown products, fibrinogen, and D-dimers 1, and 2 have been seen in infected COVID-19 patients, who also exhibit inflammation and a prothrombotic condition. A systemic inflammatory response spurred on by SARS-CoV-2 infection contributes to the generation of cytokines such as interferons and interleukins, which eventually results in the so-called cytokine storm. Cytokine concentrations at a higher amount can lead to systemic thrombus formation, with consequences of pulmonary artery thrombosis, cerebral infarction, myocardial infarction, and lower limb arterial thrombosis.24
Vaccination
The Covaxin, Covieshield, and Sputnik V vaccines, which are available in India, are well worth the investment because they effectively combat the infection. The government of India formulates the vaccination guidelines. The vaccination drive for the control of COVID-19 is in progress.
CONCLUSION
The emergence and evolution of the novel coronavirus will provide us with the knowledge we need to recognize viruses with the potential to spread widely. The current review has provided information on the structure, life cycle, variations, and strategic management employed by the Indian government to combat the Novel Corona Virus. Multiple facets of viral entry and propagation will continue to be studied in coronavirus research in the future. A better understanding of these viruses’ propensity to infect new hosts, mutation between species, and identifying substantial coronavirus reservoirs will significantly improve our abilities to anticipate when and where future epidemics may emerge.
References
- Tang X, Wu C, Li X, Song Y, Yao X, Wu X, et al. J. On the origin and continuing evolution of SARS-CoV-2. Nat Sci Rev. 2020;7(6):1012-23. [PubMed] | [CrossRef] | [Google Scholar]

- Sallard E, Halloy J, Casane D, Decroly E, van Helden J.. Tracing the origins of SARS-CoV-2 in coronavirus phylogenies: a review. Envi Chemi Lett. 2021:1-77769-85 [PubMed] | [CrossRef] | [Google Scholar]

- Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. 2020;26(4):450-2. [PubMed] | [CrossRef] | [Google Scholar]

- Vijaykrishna D, Smith GJ, Zhang JX, Peiris JS, Chen H, Guan Y., et al. Evolutionary insights into the ecology of coronaviruses. Journal of vVirology. 2007;81(8):4012-20. [PubMed] | [CrossRef] | [Google Scholar]

- Zhu Z, Lian X, Su X, Wu W, Marraro GA, Zeng Y., et al. From SARS and MERS to COVID-19: a brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respiratory research. 2020;21(1):1-4. [PubMed] | [CrossRef] | [Google Scholar]

- V’kovski P, Kratzel A, Steiner S, Stalder H, Thiel V.. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19(3):155-70. [PubMed] | [CrossRef] | [Google Scholar]

- Vijaykrishna D, Smith GJ, Zhang JX, Peiris JS, Chen H, Guan Y., et al. Evolutionary insights into the ecology of coronaviruses. Journal of vVirology. 2007;81(8):4012-20. [PubMed] | [CrossRef] | [Google Scholar]

- Krishnamoorthy S, Swain B, Verma RS, Gunthe SS. SARS-CoV, MERS-CoV, and 2019-nCoV viruses: an overview of origin, evolution, and genetic variations. VirusDisease. 2020;31(4):411-23. [PubMed] | [CrossRef] | [Google Scholar]

- Fehr AR, Perlman S.. Coronaviruses: an overview of their replication and pathogenesis. In: Coronaviruses. 2015:1-23. [PubMed] | [CrossRef] | [Google Scholar]

- Wang M-Y, Zhao R, Gao L-J, Gao XF, Wang DP, Cao JM, et al. SARS-CoV-2: structure, biology, and structure-based therapeutics development. Front Cell Infect Microbiol. 2020;10:587269 [PubMed] | [CrossRef] | [Google Scholar]

- Guruprasad L., Human SARS Human. CoV‐2 spike protein mutations. Proteins: Structure, Function, and Bioinformatics. 2021;89(5):569-76. [PubMed] | [CrossRef] | [Google Scholar]

- Cueno ME, Imai K.. Structural comparison of the SARS CoV 2 spike protein relative to other human-infecting coronaviruses. Frontiers in Medicine (Lausanne). 2020;7:1089594439 [PubMed] | [CrossRef] | [Google Scholar]

- Srivastava S, Banu S, Singh P, Sowpati DT, Mishra RK. SARS-CoV-2 genomics: an Indian perspective on sequencing viral variants. J Biosci. 2021;46(1):1-4. [PubMed] | [CrossRef] | [Google Scholar]

- Zhu Z, Lian X, Su X, Wu W, Marraro GA, Zeng Y., et al. From SARS and MERS to COVID-19: a brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respir Res. 2020;21(1):1-4224. [PubMed] | [CrossRef] | [Google Scholar]

- Ozono Seiya, Zhang Yanzhao, Ode Hirotaka, Sano Kaori, Tan Toong Seng, Imai Kazuo, Miyoshi Kazuyasu, Kishigami Satoshi, Ueno Takamasa, Iwatani Yasumasa, Suzuki Tadaki, Tokunaga Kenzo, et al. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat Commun. 2021;12:848 [PubMed] | [CrossRef] | [Google Scholar]

- Alai S, Gujar N, Joshi M, Gautam M, Gairola S.. Pan-India novel coronavirus SARS-CoV-2 genomics and global diversity analysis in spike protein. Heliyon. 2021;7(3):06564 [PubMed] | [CrossRef] | [Google Scholar]

- Sharma V, Rai H, Gautam DNS, Prajapati PK, Sharma R.. Emerging evidence on Omicron (B.1.1.529) SARS-CoV-2 variant (B. 1.1. 529) SARS‐CoV‐2 variant. J Med Virol. 2022;94(5):1876-85. [PubMed] | [CrossRef] | [Google Scholar]

- Hui DS, Chan MC, Wu AK, Ng PC. Severe acute respiratory syndrome (SARS): epidemiology and clinical features. Postgrad Med J. 2004;80(945):373-81. [PubMed] | [CrossRef] | [Google Scholar]

- Cherry JD, Krogstad P., Sars KP. The first pandemic of the 21st century. Pediatr Res. 2004;56(1):1-5. [PubMed] | [CrossRef] | [Google Scholar]

- Brooks JT, Butler JC. Effectiveness of mask wearing to control community spread of SARS-CoV-2. JAMA. 2021;325(10):998-9. [PubMed] | [CrossRef] | [Google Scholar]

- Patel A, Patel S, Fulzele P, Mohod S, Chhabra KG. Quarantine an effective mode for control of the spread of COVID19? A review. J Fam Med Prim Care. 2020;9(8):3867-71. [PubMed] | [CrossRef] | [Google Scholar]

- Parasher A.. COVID-19: current understanding of its pathophysiology, clinical presentation and treatment. Postgrad Med J. 2021;97(1147):312-20. [PubMed] | [CrossRef] | [Google Scholar]

- Tsang HF, Chan LWC, Cho WCS, Yu ACS, Yim AKY, Chan AKC, et al. An update on COVID-19 pandemic: the epidemiology, pathogenesis, prevention and treatment strategies. Expert Rev Anti-Infect Ther. 2021;19(7):877-88. [PubMed] | [CrossRef] | [Google Scholar]

- Komiyama M, Hasegawa K.. Anticoagulant therapy for patients with coronavirus disease 2019: urgent need for enhanced awareness. Eur Cardiol. 2020;15:58 [PubMed] | [CrossRef] | [Google Scholar]

