Author Notes
Copyright and License information
Citation Output
Article Metrics
Total Views
01.Kolhe P, Godge R, Ghorpade H, Dhanvate G, Nalawade A. Exploring the Anti-bacterial Potential of Novel 2-Aminophenyl-2-(2,4,5-Triphenylimidazole) Acetate Derivatives: A Comprehensive Design and Synthesis Approach. International Journal of Pharmaceutical Investigation [Internet]. 2023 Dec 27;14(1):89–100. Available from: http://dx.doi.org/10.5530/ijpi.14.1.12
ABSTRACT
Background
Antibiotic resistance is a growing concern, and the development of new anti-bacterial agents is crucial. 2-aminophenyl-2-(2,4,5-triphenylimidazole) derivatives have shown potential as anti-bacterial agents in previous studies, and this study aims to further explore their potential.Materials and Methods
Several 2-aminophenyl-2-(2,4,5-triphenylimidazole) derivatives have been developed and synthesized in this work. Using the disc diffusion technique, their anti-bacterial activity was assessed against Escherichia coli and Staphylococcus aureus. Additionally, the compounds’ drug likeness and ADMET (Absorption, Distribution, Metabolism, Excretion, and Toxicity) characteristics were assessed. To learn more about how chemical compounds attach to the biotin protein ligase, molecular docking investigations were carried out.Results
The synthesized compounds exhibited varying degrees of anti-bacterial activity, with AC6 showing the highest activity against both E. coli and S. aureus. The compounds were found to adhere to Lipinski’s rule of five, indicating good drug likeness, and exhibited favourable ADMET properties. The molecular docking studies revealed that the compounds had favourable binding modes with biotin protein ligase (PDB ID: 4DQ2).Conclusion
The 2-aminophenyl-2- (2,4,5-triphenylimidazole) derivatives designed and synthesized in this study exhibited promising anti-bacterial activity against E. coliand S. aureus. The compounds also demonstrated good Drug likeness and favourable ADMET properties. The molecular docking studies provided insights into the binding modes of the compounds with biotin protein ligase. These results suggest that 2-aminophenyl-2-(2,4,5-triphenylimidazole) derivatives have potential as anti-bacterial agents and warrant further investigation.INTRODUCTION
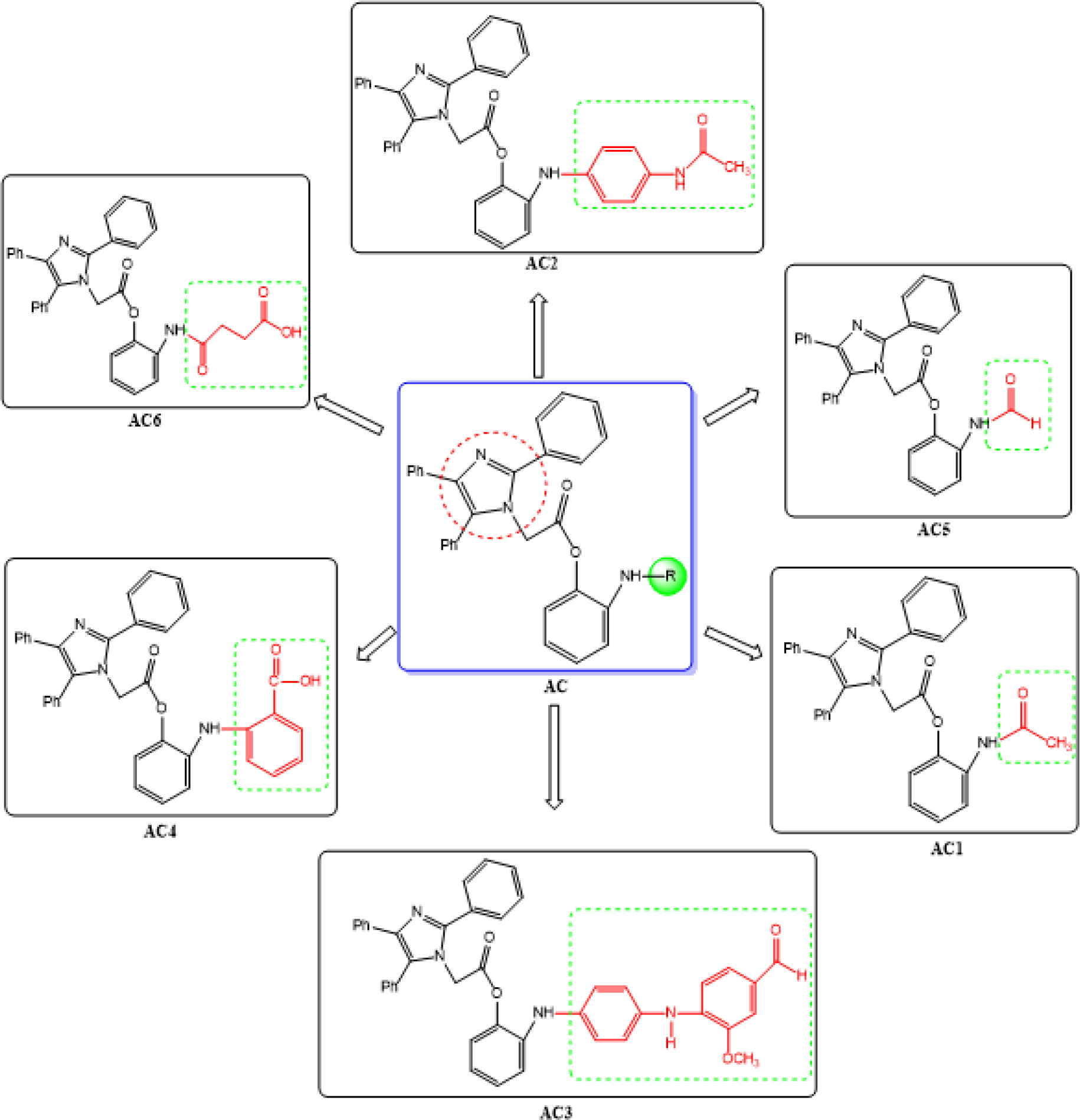
Figure 1: Designed for derivatives 2-aminophenyl-2-(2,4,5-triphenylimidazole) acetate
MATERIALS AND METHODS
Materials
Methods
ADME Parameter Estimation
Molecular Docking
Biotin Protein Ligase as a Target
In vitro Anti-bacterial Activity
RESULTS AND DISCUSSION
Chemistry and Structure-Activity Relationship (SAR)
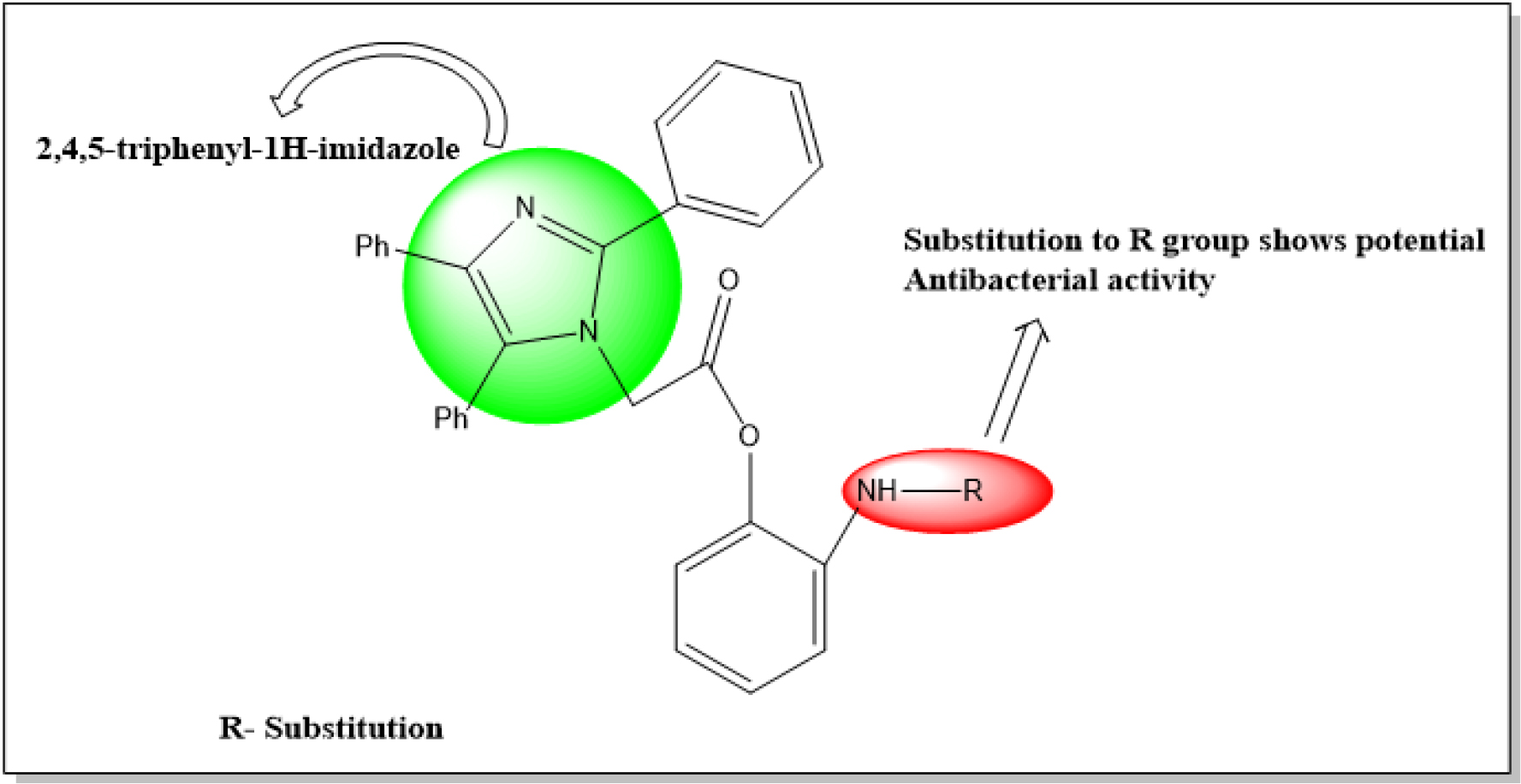
Figure 2: Design of 2-Aminophenyl-2-(2,4,5-triphenyl-1H-imidazole-1-yl)acetate derivatives.
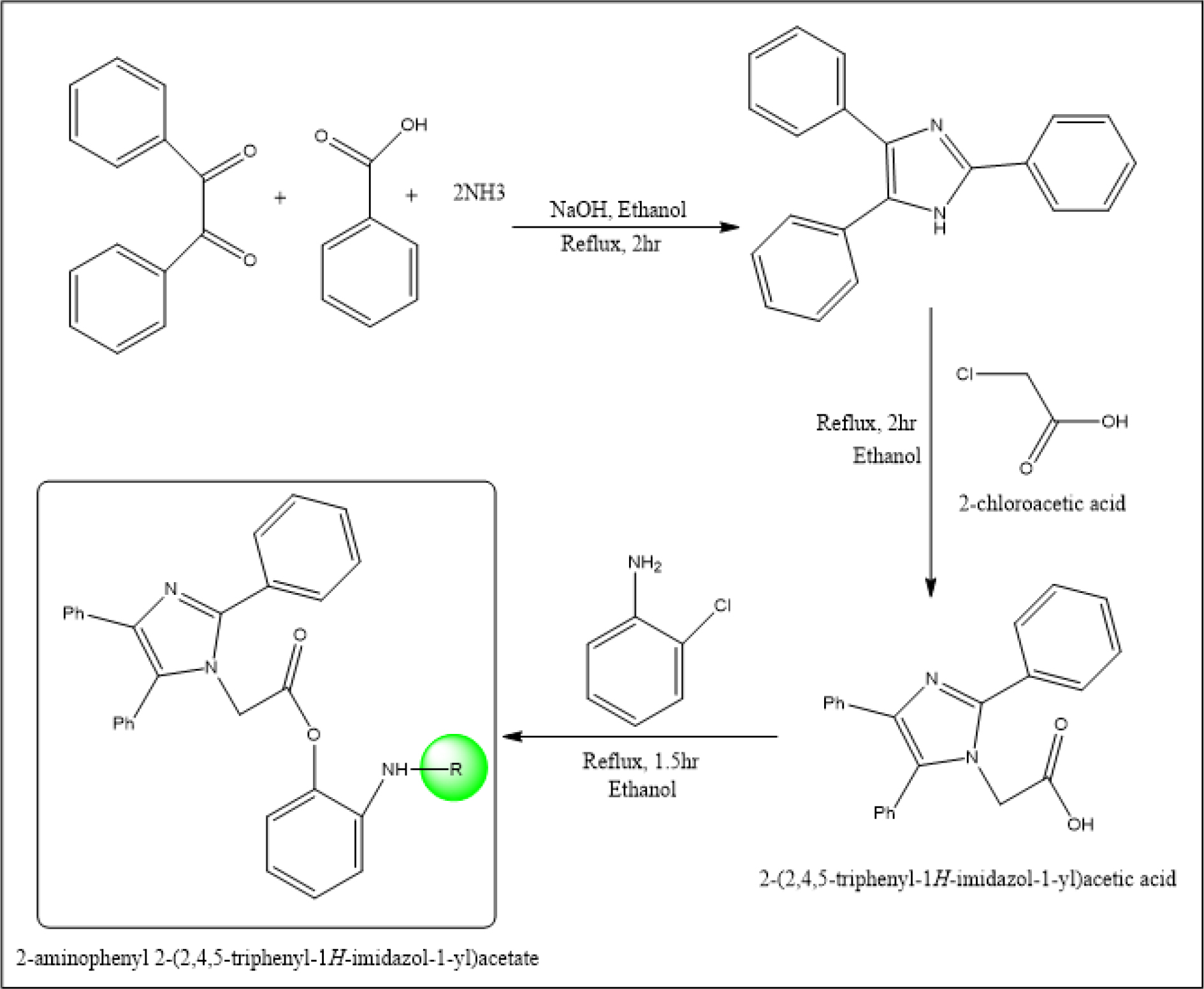
Figure 3: Scheme for synthesis of 2-Aminophenyl-2-(2,4,5-triphenyl-1H-imidazole-1-yl)derivatives (AC1 to AC6).
Synthesis and Spectral analysis of compounds (AC1-AC6)
General Procedure for synthesis of 2-amisnophenyl2- (2,4,5-triphenyl-1H-imidazole-1-yl)acetate
Synthesis of 2-acetamidophenyl 2-(2,4,5-triph enyl-1H-imidazol-1-yl)acetate (AC1)
Synthesis of 2-((4-acetamidophenyl)amino)phenyl 2-(2,4,5-triphenyl-1H-imidazol-1-yl)acetate (AC2)
Synthesis of 2-((4-((4-formyl-2-methoxyphenyl) amino)phenyl)amino)phenyl 2-(2,4,5-triph enyl-1H-imidazol-1-yl)acetate (AC3)
Synthesis of 2-((2-(2-(2,4,5-triphenyl-1-imidazol-1-yl) acetoxy)phenyl)amino)benzoic acid (AC4)
Synthesis of 2-formamidophenyl 2-(2,4,5-triph enyl-1H-imidazol-1-yl)acetate (AC5)
| Comp. | Molecular weight (g/ mol) | CMC rule violation | Lipinski’s rule violation | Mol Log P | H bond donor | H bond acceptor | No. of rotatable bonds | TPSA (Å2) |
|---|---|---|---|---|---|---|---|---|
| AC1 | 479.96 g/ mol | 2 | Yes | 4.57 | 1 | 3 | 7 | 70.14 Å2 |
| AC2 | 459.54 g/ mol | 2 | Yes | 4.30 | 1 | 3 | 7 | 70.14 Å2 |
| AC3 | 473.56 g/ mol | 2 | Yes | 4.50 | 1 | 3 | 8 | 70.14 Å2 |
| AC4 | 524.41 g/ mol | 2 | No | 4.67 | 1 | 3 | 7 | 70.14 Å2 |
| AC5 | 496.58 g/ mol | 3 | Yes | 4.74 | 1 | 3 | 7 | 70.14 Å2 |
| AC6 | 470.52 g/ mol | 2 | Yes | 3.43 | 1 | 4 | 7 | 93.93 Å2 |
| Comp. | Absorption | Distribution | Metabolism | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CaCO2 permeability (log Papp in10-6 cm/s) | GI absorption | BBB perm. (logBB) | BBB Permeant | PPB (%) | CYP3A4 substrate | CYP1A2 inhibitor | CYP2C9 inhibitor, | CYP3A4 inhibitor | CYP2C19 inhibitor | |
| AC1 | 36.94 | Low | 1.32768 | No | 100 | Weakly | No | No | Yes | Yes |
| AC2 | 47.3863 | High | 1.52328 | No | 92.600377 | Weakly | No | No | Yes | Yes |
| AC3 | 49.1514 | High | 0.842045 | No | 91.675444 | Weakly | No | No | Yes | Yes |
| AC4 | 36.0737 | Low | 1.26909 | No | 100 | Weakly | No | No | Yes | Yes |
| AC5 | 50.8426 | High | 0.523036 | No | 95.107470 | Weakly | No | No | Yes | Yes |
| AC6 | 27.1673 | High | 2.04613 | No | 93.919117 | Weakly | No | No | Yes | Yes |
Synthesis of 4-oxo-4-((2-(2- (2,4,5-triphenyl-1H-imidazol-1yl)acetoxy)phenyl) amino)butanoic acid (AC6)
Calculated Lipinski’s rule of five, drug-likeness properties and in silico ADMET analysis
Results of Molecular Docking
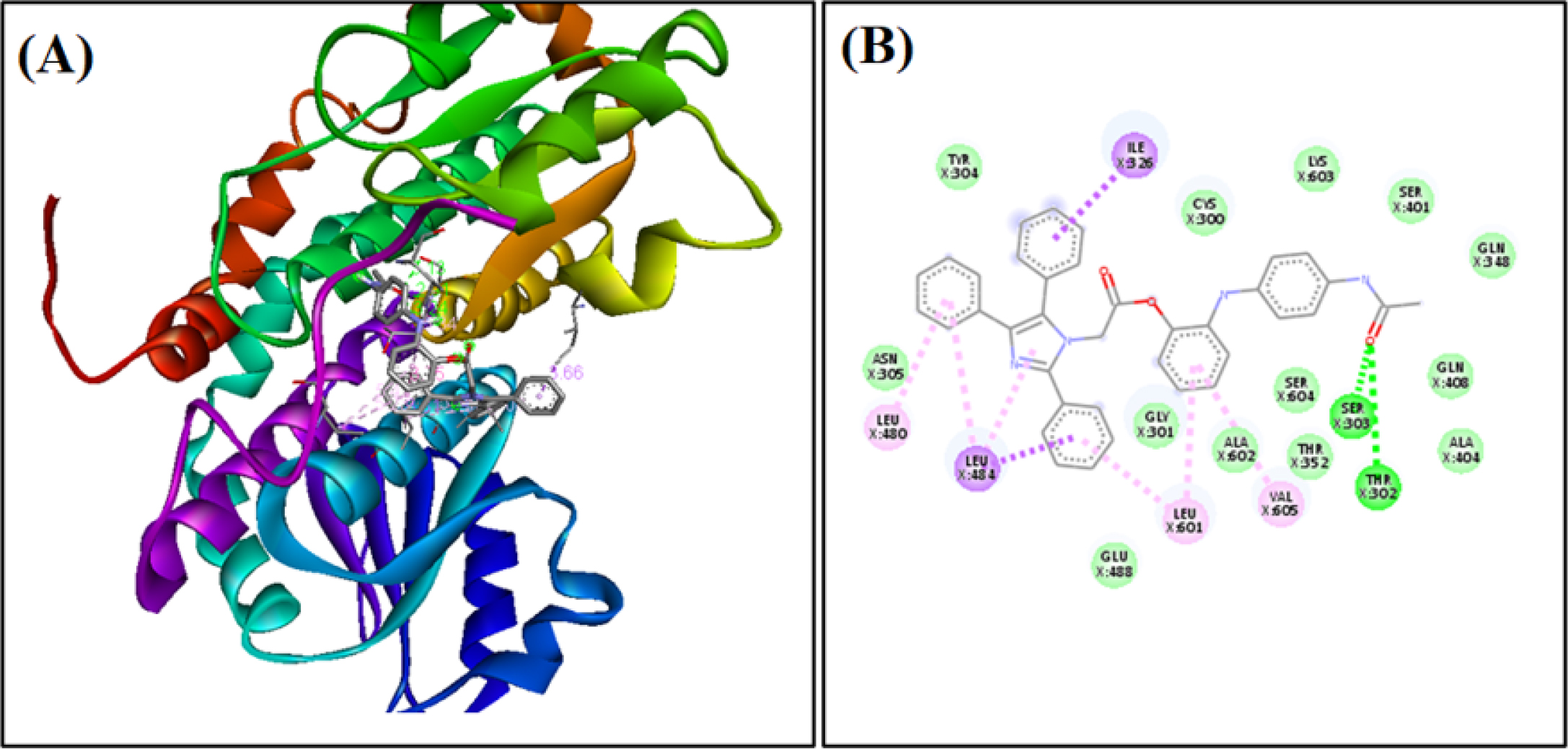
Figure 4: Binding of AC2 to active site of Biotin Protein Ligase (A) along with the 2D binding diagram (B). The nucleophilic residue is labelled in green colour and the hydrogen bonds are depicted as green dotted lines.
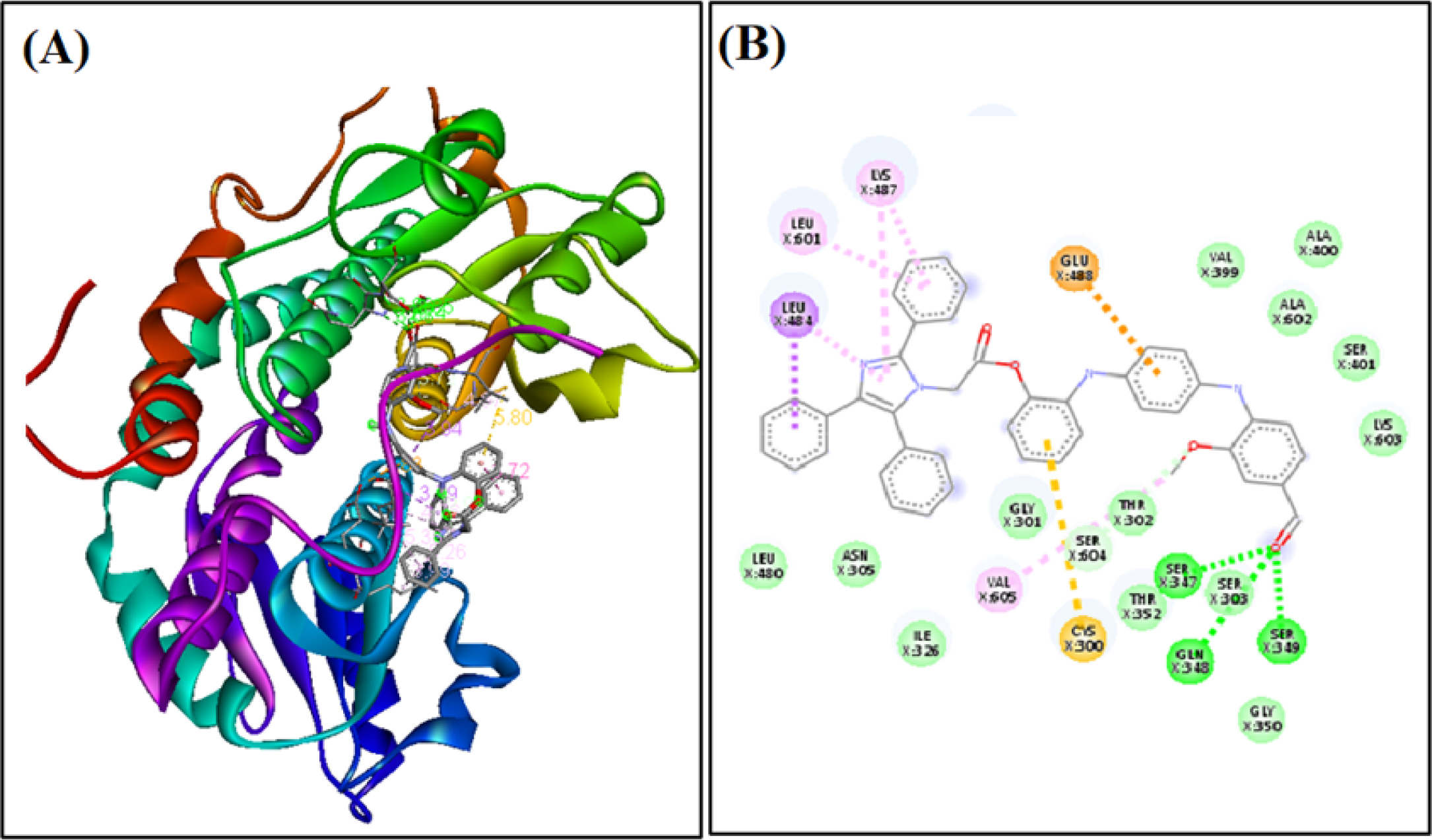
Figure 5: Binding of AC4 to active site of Biotin Protein Ligase (A) along with the 2D binding diagram (B). The nucleophilic residue is labelled in green colour and the hydrogen bonds are depicted as green dotted lines.
Results of Anti-bacterial Activity
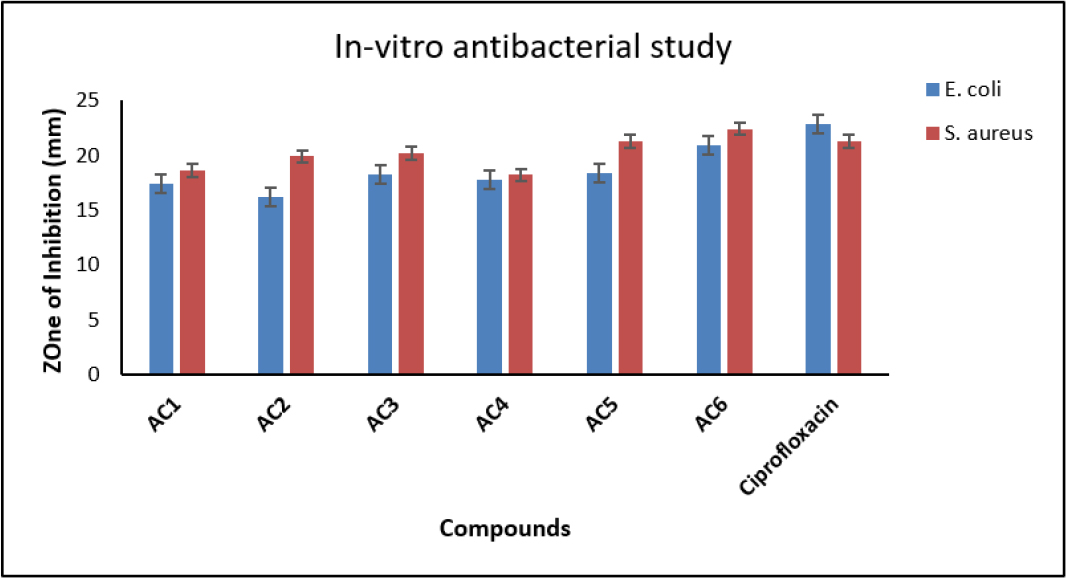
Figure 6: In vitro anti-bacterial study of synthesized compounds (AC1 to AC6).
| Compound | Active Amino Residues | Docking Score |
|---|---|---|
| AC1 | SER347, THR352, VAL605, CYS300, ASP354 | -8.9 |
| AC2 | THR302, SER303, ILE326, LEU484, UNL1, LEU601. VAL605, LEU480 | -9.4 |
| AC3 | SER347, GLN348, SER349, SER604, GLU488, UNL1, LEU484, VAL605 | -9.2 |
| AC4 | THR302, SER303, LYS487, GLU488, LEU484, LEU601, CYS300, ALA483 | -9.4 |
| AC5 | CYS300, THR352, VAL605, GLY301, ASP354, GLU488, UNL1 | -8.9 |
| AC6 | SER401, LYS603, ASP354, VAL605, UNL1, CYS300, LEU601, ILE326 | -8.1 |
| NL | GLN348, SER349, THR352, SER303, LEU601, CYS300 | -9.1 |
| Compound ID | Compounds structure | Zone of Inhibition (mm) | |
|---|---|---|---|
| E. coli | S. aureus | ||
| AC1 | 17.4±0.3 | 18.6±0.4 | |
| AC2 | 16.2±0.5 | 19.9±0.7 | |
| AC3 | 18.3±0.2 | 20.2±0.6 | |
| AC4 | 17.8±0.3 | 18.2±0.5 | |
| AC5 | 18.4±0.4 | 21.3±0.5 | |
| AC6 | 2O.9±O.3 | 22.4±0.5 | |
| Ciprofloxacin | 22.8±0.8 | 21.3±0.9 |
CONCLUSION
Cite this article
ACKNOWLEDGEMENT
ABBREVIATIONS
| ADMET | Absorption, Distribution, Metabolism, Excretion, and Toxicity |
|---|---|
| BPL | Biotin Protein Ligase |
| Caco-2 | Colorectal adenocarcinoma cell line 2 |
| DCC | Dicyclohexylcarbodiimide |
| DMAP | 4-dimethylaminopyridine |
| DMSO | Dimethyl sulfoxide |
| EI | Electron Ionization |
| ESI | Electrospray Ionization |
| FTIR | Fourier Transform Infrared Spectroscopy |
| GI | Gastrointestinal |
| HPLC | High-Performance Liquid Chromatography |
| LC-MS | Liquid Chromatography-Mass Spectrometry |
| MDR | Multidrug-resistant |
| MIC | Minimum Inhibitory Concentration |
| MBC | Minimum Bactericidal Concentration |
| MRSA | Methicillin-resistant Staphylococcus aureus |
| NMR | Nuclear Magnetic Resonance |
| PDB | Protein Data Bank |
| PPB | Plasma Protein Binding |
| SD | Standard Deviation |
| TB | Typhoid fever |
References
- Frieri M, Kumar K, Boutin A. Antibiotic resistance. J Infect Public Health. 2017;10(4):369-78. [PubMed] | [CrossRef] | [Google Scholar]
- Munita JM, Arias CA. Mechanisms of antibiotic resistance. Microbiol Spectr. 2016;4(2):481-511. [PubMed] | [CrossRef] | [Google Scholar]
- McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013;57(7):3348-57. [PubMed] | [CrossRef] | [Google Scholar]
- Marzouk AA, Abbasov VM, Talybov AH, Mohamed SK. Synthesis of 2, 4, 5-triphenyl imidazole derivatives using diethyl ammonium hydrogen phosphate as green, fast and reusable catalyst. World J Org Chem. 2013;1(1):6-10. [PubMed] | [CrossRef] | [Google Scholar]
- Yasodha A, Sivakumar A, Arunachalam G, Puratchikody A. Synthesis and biological evaluation of some 2, 4, 5-triphenyl imidazole derivatives. J Pharm Sci Res. 2009;1(4):127-30. [PubMed] | [CrossRef] | [Google Scholar]
- Rajaraman D, Sundararajan G, Rajkumar R, Bharanidharan S, Krishnasamy K. Synthesis, crystal structure investigation, DFT studies and DPPH radical scavenging activity of 1-(furan-2-ylmethyl)-2, 4, 5-triphenyl-1H-imidazole derivatives. J Mol Struct. 2016;1108:698-707. [CrossRef] | [Google Scholar]
- Jagadishbabu N, Shivashankar K. One-pot synthesis of 2, 4, 5-triphenyl imidazoles from 1, 2-diols as key reagents. J Chin Chem Soc. 2017;64(5):474-80. [CrossRef] | [Google Scholar]
- Boyatzis S, Nikokavouras J. Absorption, fluorescence and chemiluminescence spectra of 2,4,5-Triphenylimidazole (lophine) and 2-(p-dimethyl-aminophenyl)-4,5-Diphenylimidazole in micellar solutions. Mol Cryst Liq Cryst. 1986;137(1):403-12. [CrossRef] | [Google Scholar]
- Jayachandran JP, Wang ML. N-alkylation of 2, 4, 5-triphenyl imidazole derivatives using a new phase transfer reagent under PTC conditions. Synth Commun. 2001;31(18):2743-52. [CrossRef] | [Google Scholar]
- Vijesh AM, Isloor AM, Telkar S, Arulmoli T, Fun HK. Molecular docking studies of some new imidazole derivatives for antimicrobial properties. Arab J Chem. 2013;6(2):197-204. [CrossRef] | [Google Scholar]
- Norinder U, Bergström CA. Prediction of ADMET properties. ChemMedChem. 2006;1(9):920-37. [PubMed] | [CrossRef] | [Google Scholar]
- Sakaino Y, Kakisawa H, Kusumi T, Maeda K. Structures and chromotropic properties of 1, 4-bis(4, 5-diphenylimidazol-2-yl)benzene derivatives. J Org Chem. 1979;44(8):1241-4. [CrossRef] | [Google Scholar]
- Vijesh AM, Isloor AM, Telkar S, Peethambar SK, Rai S, Isloor N, et al. Synthesis, characterization and antimicrobial studies of some new pyrazole incorporated imidazole derivatives. Eur J Med Chem. 2011;46(8):3531-6. [PubMed] | [CrossRef] | [Google Scholar]
- Petersen PJ, Jacobus NV, Weiss WJ, Sum PE, Testa RT. In vitro and in vivo anti-bacterial activities of a novel glycylcycline, the 9-t-butylglycylamido derivative of minocycline (GAR-936). Antimicrob Agents Chemother. 1999;43(4):738-44. [PubMed] | [CrossRef] | [Google Scholar]
- Tanino H, Kondo T, Okada K, Goto T. Structures of three isomeric dimers of 2,4,5-Triphenylimidazolyl. Bull Chem Soc Jpn. 1972;45(5):1474-80. [CrossRef] | [Google Scholar]
- Gazieva GA, Karpova TB, Nechaeva TV, Kravchenko AN. Ring contraction of 1,2,4-triazine derivatives in the synthesis of imidazoles. Russ Chem Bull. 2016;65(9):2172-82. [CrossRef] | [Google Scholar]
- Liu C, Shi C, Mao F, Xu Y, Liu J, Wei B, et al. Discovery of new imidazole derivatives containing the 2, 4-dienone motif with broad-spectrum antifungal and anti-bacterial activity. Molecules. 2014;19(10):15653-72. [PubMed] | [CrossRef] | [Google Scholar]
